Theoretical and computational biophysics
|
Statistical
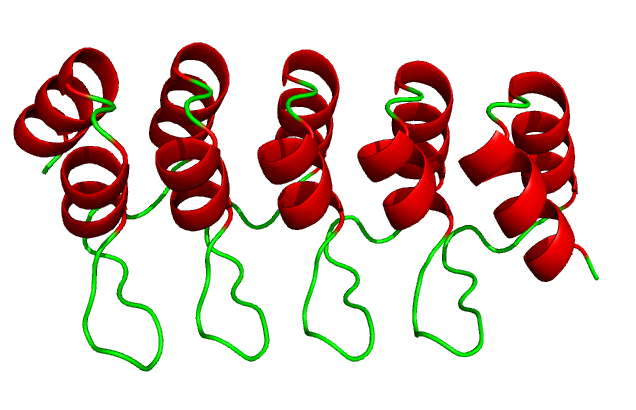
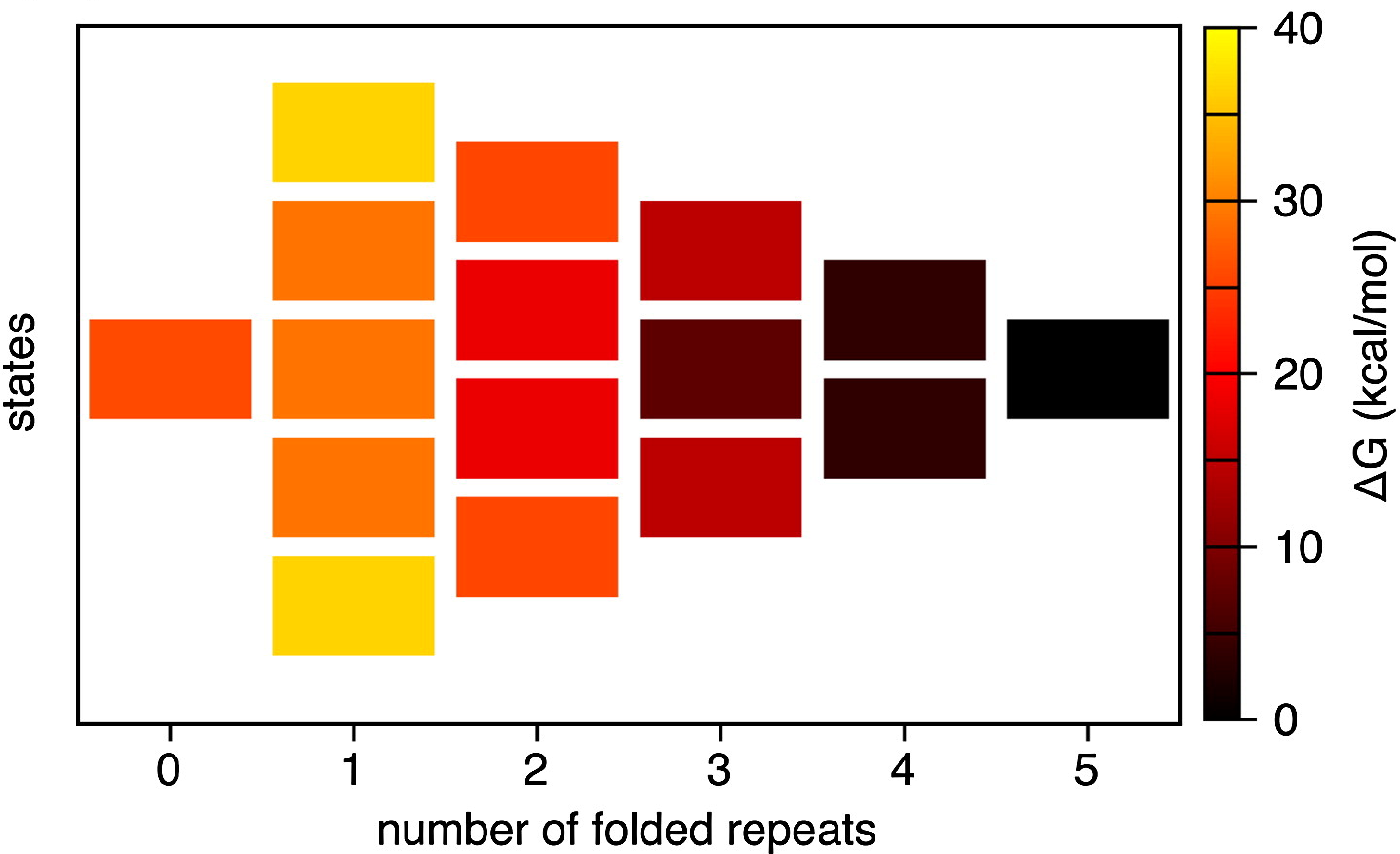
mechanical modeling and simulations of repeat
proteins. Repeat protein domains are
formed by tandem arrays of repeating structural
units, constitute about 20% of the eukaryotic
proteome, mediating protein-protein interactions
and acting as mechano-transductors. As such they
may represent the basis for the construction of
mechanical nanodevices. In collaboration with
experimental groups in the field, we have been
working on simplified models of repeat proteins
which explains both the thermodynamics and the
kinetics of folding of this class of proteins.
We have also been carrying out atomistic
molecular dynamics (MD) simulations of several
repeat protein systems to study their folding
behavior and their mechanical characteristics
when subjected to external pulling forces. References:
|
|
|
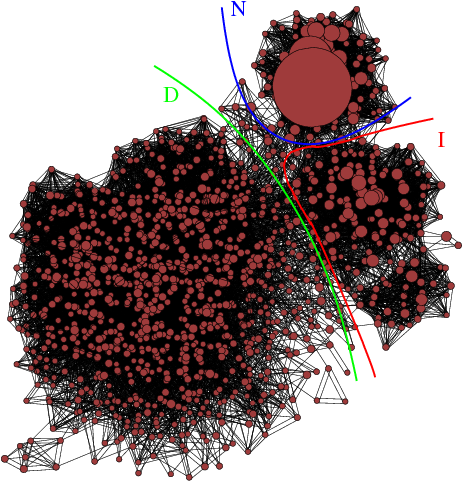
Peptide folding. Our
activity focuses on the development and
application of methods for the identification of
the folding transition state of peptides
and, more in general, for the complete
characteriztion and representation of the
dynamics of peptides by using atomistic
molecular dynamics simulations . This research
effort is based on the application of concepts
like kinetic networks and Markov models to the
trajectory data of peptides collected by MD
simulations. Results from this line of research
are validated against available experimental
data on the kinetics of folding of peptides
(folding/unfolding rates, phi values). References:
|
|
|
Transport and adsorption properties of blood proteins. Transport of nutrients to peripheral tissues and healing of damaged blood vessels are among the most important functions of blood. These functions involve the action of a series of proteins some of which are found in large amounts in the blood circulation. Fibrinogen is a multiprotein complex which, when activated, aggregate to form fibrin, a net-shaped molecular formation which is fundamental for the coagulation of blood following, i.e, a wound or when an extraneous body comes into contact with blood (i.e., graft implants). Thus, adsorption of fibrinogen on material surfaces play an important role in viability of those materials for implants. In collaboration with experimental groups in the field, we use atomistic molecular dynamics simulations to characterize the adsorption process of fibrinogen on material surfaces. Another important molecule in the blood is albumin, which mediate transport of lipids and other molecules in blood. Albumin is a multidomain protein which provides several binding sites used to bind a range of different target molecules. Target molecules (lipids, drugs, etc.) bind to albumin which act as a transporter, and are then released where needed by blood circulation. Here we use molecular dynamics simulations to study the binding modes of several lipids to Albumin and the kinetics of lipid release/uptake. Figure Caption: Simulation box containing a water solvated fibrinogen molecule on a slab of amorphus silica. |
|